Basics of Multivariate Analysis II (Principal Component Analysis): Theory and Exercise using R by Takeshi Furuhashi
Author:Takeshi Furuhashi [Takeshi Furuhashi]
Language: eng
Format: epub
Published: 2015-08-04T16:00:00+00:00
The fourth script
x_centered <- scale (x_somatometry, apply(x_somatometry, 2, mean), c(1, 1)),
will center the data in each column in Table 2.2 (see Sec. 2.3 ). The term mean refers to an average, apply (a, 2, mean) is the R function to calculate the mean value of each column in matrix a, while 2 indicates a column (if this is 1, then a row is indicated instead). The output of the apply() function in this case is a two-dimensional (2D) vector. c(p, q) is also a 2Dl vector. IF p = q = 1, then each element of the vector is 1. The function scale(x, mean, v) extracts the mean from x, and divides x by v. In the case of Table 2.2, x is a 20 × 2 matrix, and the mean and v are 2 × 1 vectors. From each element in the first column of x, the value in the first row of the mean is subtracted, and the resulting value is divided by the value in the first row of v. Because the value in the first row of c(1, 1) is 1, this does not change the value of the dividend.
By executing the plot() function below
plot(x_centered[,1],x_centered[,2],col=“red”, pch=1),
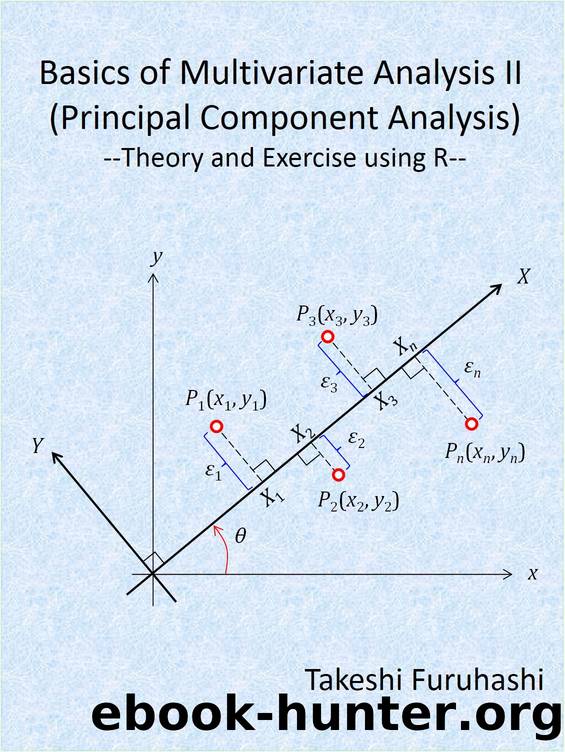
the distribution in Fig.2.10 (see Subsec.2.3.1 ) is obtained. After this data centering is complete, the scripts for the PC analysis are the same as those in Sec.2.2 . Fig. 2.13 shows the identified PC axis.
Download
This site does not store any files on its server. We only index and link to content provided by other sites. Please contact the content providers to delete copyright contents if any and email us, we'll remove relevant links or contents immediately.
Secrets of the JavaScript Ninja by John Resig Bear Bibeault(5955)
Linux Device Driver Development Cookbook by Rodolfo Giometti(3406)
Implementing Enterprise Observability for Success by Manisha Agrawal and Karun Krishnannair(3036)
TCP IP by Todd Lammle(2637)
Drawing Shortcuts: Developing Quick Drawing Skills Using Today's Technology by Leggitt Jim(2531)
Pandas Cookbook by Theodore Petrou(2498)
Applied Predictive Modeling by Max Kuhn & Kjell Johnson(2478)
Supercharging Productivity with Trello by Brittany Joiner(2290)
Design Made Easy with Inkscape by Christopher Rogers(2211)
Learn Qt 5: Build modern, responsive cross-platform desktop applications with Qt, C++, and QML by Nicholas Sherriff(2175)
40 Algorithms Every Programmer Should Know by Imran Ahmad(2146)
Mastering Tableau 2023 - Fourth Edition by Marleen Meier(2009)
Fusion 360 for Makers by Lydia Sloan Cline(1985)
Build Stunning Real-time VFX with Unreal Engine 5 by Hrishikesh Andurlekar(1985)
Inkscape by Example by István Szép(1873)
The Artificial Intelligence Imperative by Anastassia Lauterbach(1845)
Customizing Microsoft Teams by Gopi Kondameda(1806)
The Old New Thing by Raymond Chen(1719)
Mastering Python Scientific Computing by Hemant Kumar Mehta(1707)
